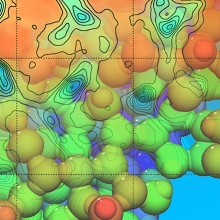
Machine-learned potentials have led to a real breakthrough in theoretical chemistry in recent years. The potential energy surface is approximated by a neural network rather than calculated at each geometry using DFT. We can now simulate slow and rare processes that include the breaking and forming of chemical bonds, like adsorption or reflection of particles on a surface, phase transitions, or dislocations, by ML-based accelerated molecular dynamics calculations.
We developed Gaussian moments [1,2], a molecular descriptor that encodes atomistic geometries for neural networks. As a physically-informed descriptor, it is invariant to rotation, translation, and the exchange of like atoms (e.g., two hydrogen atoms). We can easily tune it between accuracy and efficiency.
Active learning techniques ensure data efficiency. Using a sophisticated error measure for neural networks [3], we perform batch active learning [4] to make sure we need as few DFT training data as possible while still maintaining accuracy.
Our developments are available as open-source codes. Our pilot implementation, GM-NN, is focused on active learning. A re-implementation, Apax, focuses on performance and user- and developer-friendliness. It enables learning on the fly with IPSuite.
We have applied our methods to surface astrochemistry [5], the diffusion of atoms on a surface [6], and the ML prediction of magnetic properties (anisotropy tensors), which we use to predict spin-phonon coupling [7].
In an alternative approach, we use Gaussian-process regression (GPR) to accelerate geometry optimizations and transition-state search in a learning-on-the-fly manner. Reaction-path searches can be handled very efficiently and stable using GPR. All these developments are available in DL-FIND.
Are you interested in working with us? We have found that many students who are fascinated by ML initially learn that their daily work is somewhat focused on programming and algorithms. We are happy to train you in ML, and its use in chemistry, but any previous experience in that area certainly helps.